CAREER: NetDA—Protein Network-Based Software for Disease Analysis Using Cliques, Bipartite Graphs, and Diffusion Kernels
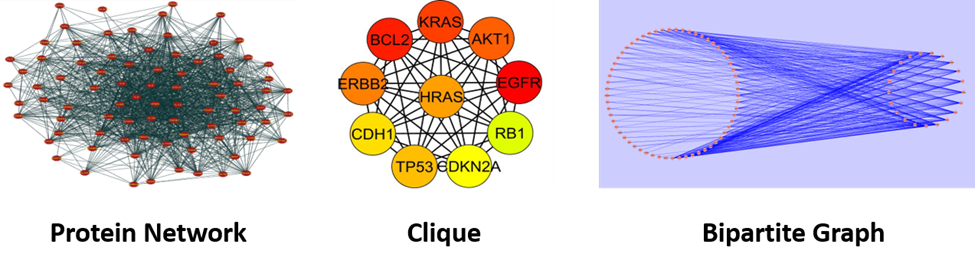
The primary objective of this research is to investigate and analyze the complex phenomena of disease progression at the protein network level using three graph theoretic concepts – clique, bipartite graph, and diffusion kernel. Computational studies of disease progression are needed since experimental studies are expensive and involve sacrificing animals like mice and rats. For humans, samples are collected during surgery of patients at different stages of a disease. At each stage, this protocol requires multiple patients to have statistically significant outcomes, which makes experimental study complicated. With humans, experimental studies also raise significant ethical and socio-cultural issues. Thus, computational analysis is needed to understand the essential mechanisms of disease progression as transitions between disease stages in protein networks that hold the key to early diagnosis. This work will analyze disease progression as an event-schedule-like structure by developing the software toolkit NetDA (Network-based Disease Analysis), where a) each event representing a disease stage completed by a group of proteins will be analyzed in terms of clique and clique-like graphs, b) transfer of signals from one stage to the next will be analyzed using bipartite and bipartite-like graphs, and c) the strength of signals will be analyzed using diffusion kernel. The outcome of this study will help not only in early diagnosis of a disease but also in drug design specific for a disease stage.
Dates Active: October 2018 — June 2021
Organizations
National Science Foundation (NSF)
