Memory-Distributed Computing for Big Mass-Spectrometry based Omics
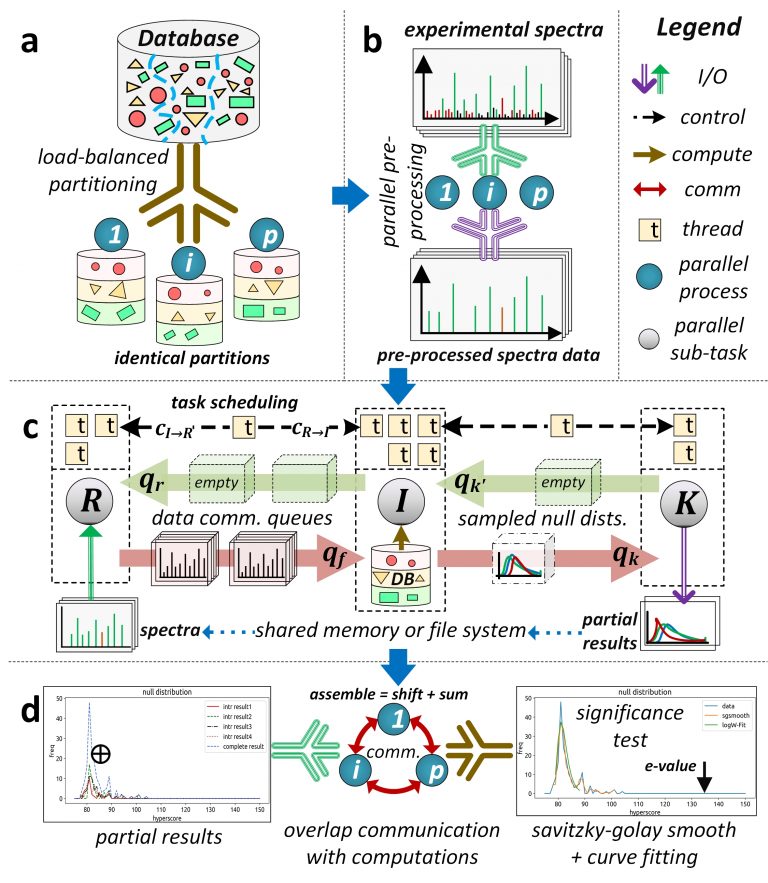
Database peptide search algorithms deduce peptides from mass spectrometry data. There has been substantial effort in improving their computational efficiency to achieve larger and more complex systems biology studies. However, modern serial and high-performance computing (HPC) algorithms exhibit suboptimal performance mainly due to their ineffective parallel designs (low resource utilization) and high overhead costs. We present an HPC framework, called HiCOPS, for efficient acceleration of the database peptide search algorithms on distributed-memory supercomputers. HiCOPS provides, on average, more than tenfold improvement in speed and superior parallel performance over several existing HPC database search software. We also formulate a mathematical model for performance analysis and optimization, and report near-optimal results for several key metrics including strong-scale efficiency, hardware utilization, load-balance, inter-process communication and I/O overheads. The core parallel design, techniques and optimizations presented in HiCOPS are search-algorithm-independent and can be extended to efficiently accelerate the existing and future algorithms and software.
Dates Active: April 2017 – April 2020
Organizations
National Institutes of Health (NIH)
